 Home / Services / In Vivo Functional Characterization / In Vivo Identification of Kinase/Phosphatase Substrates
Home / Services / In Vivo Functional Characterization / In Vivo Identification of Kinase/Phosphatase Substrates In Vivo Identification of Kinase/Phosphatase Substrates
INQUIRYCreative BioMart offers a full range of services and advanced techniques to assist our customers identify in vivo substrate of kinases and phosphatases. Our scientific team will use their unparalleled expertise and experience in kinase/phosphatase biology to provide you with the highest quality service.
Background
Phosphorylation and dephosphorylation are well-known regulatory mechanisms involved in most of the biological processes. While our understanding of kinase/phosphatase biology has improved extraordinarily, the discovery of substrates for kinases and phosphatases remains a limiting step in the elucidation of cellular signaling processes. To date, no single method has proven to be a panacea for identifying substrates.
Since potential substrates discovered in vitro must be verified in intact cells, these traditional methods of identifying kinase/phosphatase substrates in vitro are now being complemented by methods of identifying substrates in vivo. Recent advances in technologies such as proteomics have the potential to facilitate the development of new methods to identify substrates of kinases and phosphatases in vivo. These techniques have greatly advanced our understanding of the underlying mechanisms of phosphatase signaling pathways.
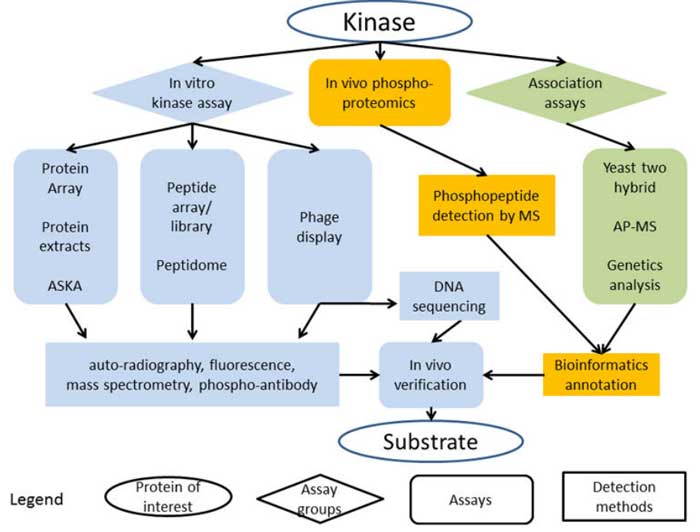
Figure 1. High throughput strategies to identify kinase substrates. (Xue L & Tao W A., 2013)
Our Services
Creative BioMart has an experienced scientific team to provide in vivo substrate identification services for a wide variety of kinases and phosphatases. Whether your purpose is to identify a substrate that interact with your enzyme of interest or to identify an enzyme that can interact with a specific substrate, we have the ability to deliver the desired results within your expected time.
Substrate Prediction
We have developed an integrative computational platform combining multiple bioinformatics tools and algorithms to predict the interaction between kinases/phosphatases and their potential substrates. Our algorithms cover almost all kinases and phosphatases. The localization of enzymes and their predicted substrates is also considered as an important parameter. We also provide follow-up experimental verification services for predicted substrates.
Enrichment of Phosphoproteins and Phosphopeptides
We have established a platform covering advanced techniques including metal oxide/hydroxide affinity chromatography (MOAC), strong cation exchange chromatography (SCX), immobilized metal affinity chromatography (IMAC), and immunoprecipitation to help our customers overcome the challenges of phosphorylation study due to the low abundance of phosphoproteins. We offer a full range of services covering sample preparation, protein extraction to protein/peptide identification to help our customers save time and effort.
Identification of In Vivo Substrates
We provide different strategies including in vivo phosphoproteomics, chemical genetics, isotope labeling, yeast two-hybrid system, and protein arrays for in vivo identification of kinase/phosphatase substrates. Our scientific team specializing in protein interaction network analysis has the ability to identify potential substrates and validate kinase/phosphatase-substrate interactions in vivo in a highly efficient manner.
Creative BioMart is an advanced biotechnology company providing comprehensive services and products to customers engaged in kinase/phosphatase biology research and drug discovery. Our scientific team will work closely with our customers to ensure that the services we provide will actually meet their expectations. If you require further information about any of our services or products, please feel free to contact us at any time. We will get in touch with you as soon as possible.
References
- Xue L & Tao W A. Current technologies to identify protein kinase substrates in high throughput. Frontiers in biology, 2013, 8(2): 216-227.
- Linding R, et al. Systematic discovery of in vivo phosphorylation networks. Cell, 2007, 129(7): 1415-1426.

